
Selected Publications
Biocatalytic Construction of a Chemical Exchange Saturation Transfer Magnetic Resonance Imaging Nucleoside Probe: Synthesis and Evaluation of 5‐Methyl‐5, 6‐Dihydrothymidine
https://advanced.onlinelibrary.wiley.com/doi/abs/10.1002/adsc.70200
A collaboration with the Gilad group at Michigan State University to develop a novel biosynthesis pathway for 5-MDHT, a potent MRI contrast agent. In this work, we used AMBER as the simulation package to predict the catalytic mechanism, in which Met219 and the sugar-phosphate αR1P sandwich the uracil analog, ensuring efficient production of the nucleotide 5-MDHT as the experimental results predict.

Cooperative and inhibitory ion transport in functionalized angstrom-scale two-dimensional channels
https://www.nature.com/articles/s41467-025-61307-x
A collaboration with the Schatz group and Northwestern University. In this work, I provided the 12-6-4 parameters for Pb2+ based on experimental HFE and DFT-calculated IOD/CN provided by the Schatz group. The parameter performs well at predicting the ion channel's ion selectivity and inhibition, demonstrating that the 12-6-4 model remains a robust method, even in the biomaterial field.

Quantum-Centric Alchemical Free Energy Calculations
https://arxiv.org/abs/2506.20825
First co-first author paper in the quantum field. In this work, I developed a Fortran-Python interface (more accurately, a Python monkey patch for Fortran) to facilitate crosstalk between QUICK, PySCF, and IBM's quantum hardware. Dr. Milana Bazayeva conducted a meticulous review of my code and performed thorough tests to calculate the relative solvation free energy between ammonia and methane using QM/MM.
Dr. Akhil Shajan and Dr. Danil Kaliakin reviewed the article, and Dr. Fangchun Liang generated awesome figures. Dr. Susanta Das adapted the same idea for QM/MM PMF.
Yes, we can utilize various free energy methods with quantum hardware.

Towards quantum-centric simulations of extended molecules: sample-based quantum diagonalization enhanced with density matrix embedding theory
https://pubs.acs.org/doi/full/10.1021/acs.jctc.5c00114
Another quantum computing paper was submitted to the Quantum Physics Archive. In this work, we extended the previous SQD method and combined it with the density matrix embedding theory (DMET) to achieve divide-and-conquer on larger molecules. We are able to accurately calculate the conformation change landscape of cyclohexane, as well as the expansion/shrinking of an H18 ring, with results comparable to the CCSD(T) level. Again, I mainly contributed to the parallelization of DMET-SQD code in this work.

Accurate quantum-centric simulations of supramolecular interactions

https://www.nature.com/articles/s42005-025-02305-9
First quantum computing paper submitted to Quantum Physics Archive. In this work, a collaboration between CCF and IBM was established to answer an essential question: Can quantum computer hardware be used to calculate hydrophilic and hydrophobic interactions? The answer is YES. Powered by the most up-to-date quantum postprocessing code, sample-based quantum diagonalization (SQD), we are able to accurately calculate the dissociation curve of hydrophilic and hydrophobic molecules comparable to the CCSD(T) level. I mainly contributed to the parallelization of SQD code in this work.
Simulating Metal-Imidazole Complexes
https://pubs.acs.org/doi/10.1021/acs.jctc.4c00581
Finally, after three years of troubleshooting, we offered a more convenient yet still accurate solution to the problem. "metal imidazole simulation tends to be inconsistent with the experiment".
Turns out that when the multiple imidazole complex forms, each imidazole plane is prone to form an angle with the coordinating line. This newly-discovered phenomenon is a result of multiple factors, including cation-pi interactions and hydrogen bonds between imidazoles.

Mg2+-Ion Dependence Revealed for a BAHD 13-O-β-Aminoacyltransferase from Taxus Plants
https://pubs.acs.org/doi/full/10.1021/jacsau.4c00577
Continuation of the collaboration with Dr. Kevin Walker. Here, Dr. Aimen Al-Hilfi from the Walker Group discovered a new baccatin III enzyme catalytic mechanism that is heavily Mg2+ dependent. I then ran MD simulations using pairwise C4 interactions to support this idea.

Binding of Phosphate Species to Ca2+ and Mg2+ in Aqueous Solution
https://pubs.acs.org/doi/10.1021/acs.jctc.4c00218
A collaboration with Dr. Viktorya Aviyente from Bugazici University in Turkey. Here, we continued using PMF, TI and QM methods to parametrize proper parameters to describe metal-phosphate interactions. The biggest challenge here is the multiple charge states of phosphate, and we have to provide parameters for each charge species. We also provided an AMBER tutorial for building up this system.

Biocatalytic and Regioselective Exchange of 2-O-Benzoyl for 2-O-(m-Substituted) BenzoylGroups to Make Precursors of Next-Generation Paclitaxel Drugs

https://chemistry-europe.onlinelibrary.wiley.com/doi/abs/10.1002/cctc.202400186
A collaboration with Dr. Kevin Walker from the MSU Chemistry Department. In this work, Gaussian calculation was used to investigate how different functional groups on C13 of the baccatin III molecule will affect the substrate conformation. Then, an MD simulation was carried out to validate the theory that a carbonyl group on C13 will make the substrate have the highest catalytic activity on C2 debenzoylation, which matches the experimental results.
Thermodynamics of Metal–Acetate Interactions

https://pubs.acs.org/doi/full/10.1021/acs.jpcb.3c06567
The metal-acetate interaction paper is finally out! Majid did a great job at discussing how the preferences on monodentate/bidentate states of the acetate are altered after using the new parameter sets (namely, the new polarizability of the oxygen on acetate). A new AMBER tutorial on how to use these parameters is on its way. Stay tuned!
Rational engineering of an elevator-type metal transporter ZIP8 reveals a conditional selectivity filter critically involved in determining substrate specificity

https://www.nature.com/articles/s42003-023-05146-w
A collaboration with Dr. Jian Hu from MSU Biochemistry and Molecular Biology Department. In this work, Thermodynamic Integration (TI) was used to varify that a 4-residue-mutated variable of human ZIP8 protein has higher selectivity of Zn2+ over Cd2+. This discovery can possibly shed light to cure human Cd2+ poisoning.
Accurate Metal–Imidazole Interactions
https://pubs.acs.org/doi/abs/10.1021/acs.jctc.2c01081
Another cut-edge research work thanks to Dr. Lin Song's innovative idea and hard working. In this article, the polarizability of imidazole nitrogen was fine-tuned to reproduce experimental binding energy between metal ions and histidines. This provided a full set of parameters to support more accurate metalloprotein simulation and closes the gap between aqueous and protein MDs.


Systematic Evaluation of Ion Diffusion and Water Exchange
https://pubs.acs.org/doi/full/10.1021/acs.jctc.1c01189
My first independent work in the Merz Group. Thanks to Dr. Merz's inspiration, this paper did a further evaluation on the quality of Li/Merz metal ion parameters by testing the parameters systematically on diffusion and water exchange. The diffusion and exchange rate protocol was also novelized with new ways of determining cutoff, shorter but still converging simulation length, as well as nested-averages to enhance sampling randomness and simulation accuracy.

A Polarizable Cationic Dummy Metal Ion Model
https://pubs.acs.org/doi/10.1021/acs.jpclett.2c01279
A collaboration with Dr. H. Metin Aktulga in MSU CSE department. Ali, Kurt and Cagri are all brilliant fellows coming up with an idea that allows cation-dummy-atom (CDA) model to have dynamic charges along the simulation frames. I contributed a little on the scanning protocol that leads to Figure 4 and 5.


Parametrization of Trivalent and Tetravalent Metal Ions for the OPC3, OPC, TIP3P-FB, and TIP4P-FB Water Models
https://pubs.acs.org/doi/10.1021/acs.jctc.0c01320
My second work as the first author. Have to say it is a bit repetitive compare to the divalent paper, but the 2D quadratic fitting model has been improved a lot for a much quicker parametrization process (please see Figure 1 of the paper for details). Also, the diffusion coefficient benchmarking interface ISAIAH has been developed with higher robustness and user friendliness. Please check my GitHub page for more information on ISAIAH.
Systematic Parametrization of Divalent Metal Ions for the OPC3, OPC, TIP3P-FB, and TIP4P-FB Water Models
https://pubs.acs.org/doi/abs/10.1021/acs.jctc.0c00194
My first work as the first author. Using 2D quadratic fitting, the perfect parameter combination for a metal ion to reproduce both HFE and IOD can be found with high precision. I also derived a protocol to calculate diffusion coefficient of metal ions for various parameter sets.
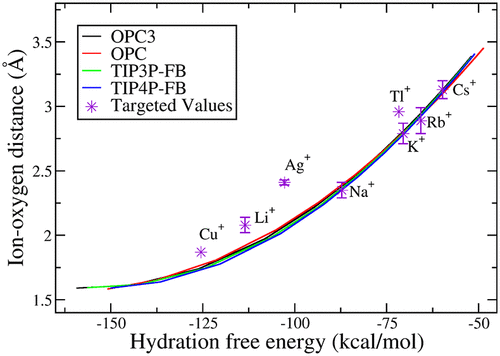
Parameterization of Monovalent Ions for the OPC3, OPC, TIP3P-FB, and TIP4P-FB Water Models
https://pubs.acs.org/doi/10.1021/acs.jcim.0c01390
Similar to the paper above, but I only participated in the halide anion part. I also used the Kirkwood-Buff integral to calculate activity derivative of common salt solutions.
A surface-display biohybrid approach to light-driven hydrogen production in air
https://advances.sciencemag.org/content/4/2/eaap9253
My very very first work as a scientist back in China. I dealt with the plasmid construction of OmpA-PbrR and HyaABCDEF using PCR and dual-antibiotics colony screening. I also participated in nano-coating of CdS particle on the surface of E.coli and the subsequent SiO2 encapsulation using PDADMAC and PSS cross-linking followed by silicic acid bath.

Proteolysis mediated by the membrane‐integrated ATP‐dependent protease FtsH has a unique nonlinear dependence on ATP hydrolysis rates
https://onlinelibrary.wiley.com/doi/full/10.1002/pro.3629
My first work in MSU, I conducted casein ATPase assay with the control set using Lon, a soluble AAA+ protease to validate the sigmoidal behavior of FtsH proteolysis over ATP concentration. Lon shows linear trend instead, as displayed in Figure 3.

Implementation of Mealy Machine with Molecular Reactions
https://ieeexplore.ieee.org/abstract/document/8422571
A paper published during my undergrads as a collaboration with Southeastern University in China. My work was using ABI real-time PCR machine to monitor the fluorescent signal intensity over time for the molecular mealy machine. DABCYL was used as a universal quencher, while TAMRA and SYBR Green are used as 5' and 3' labels.
